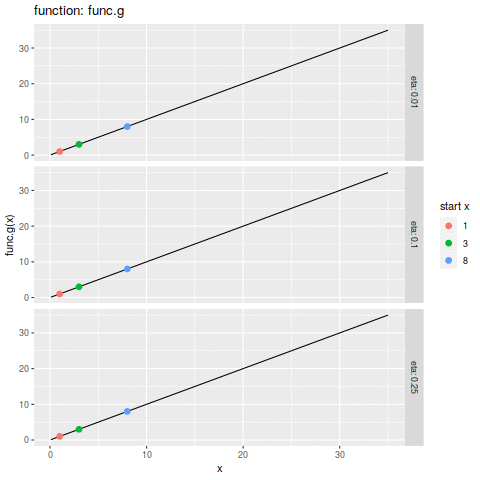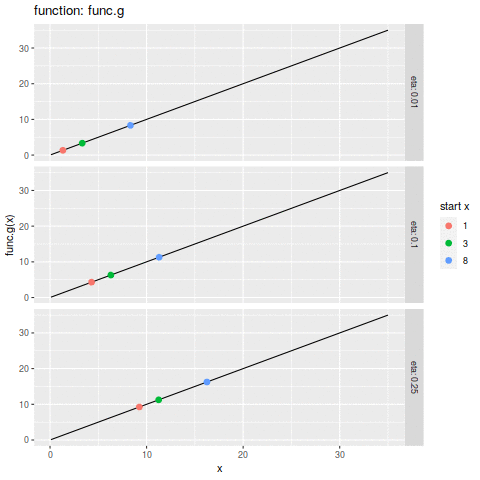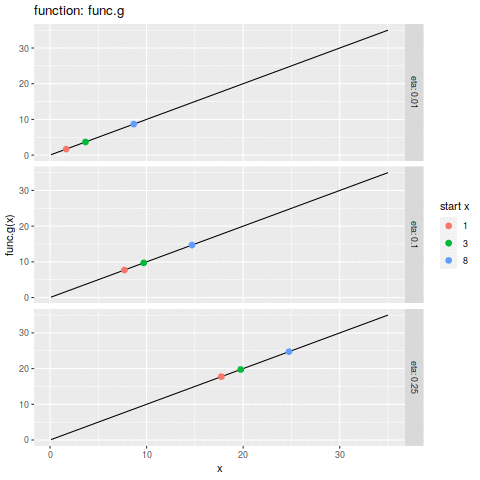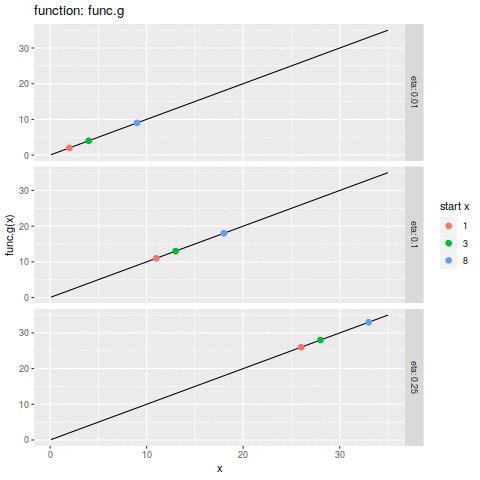
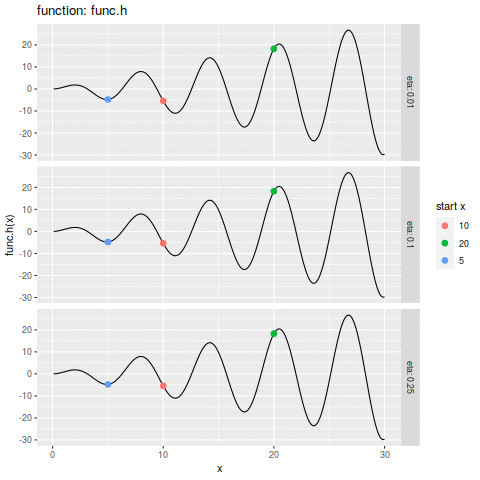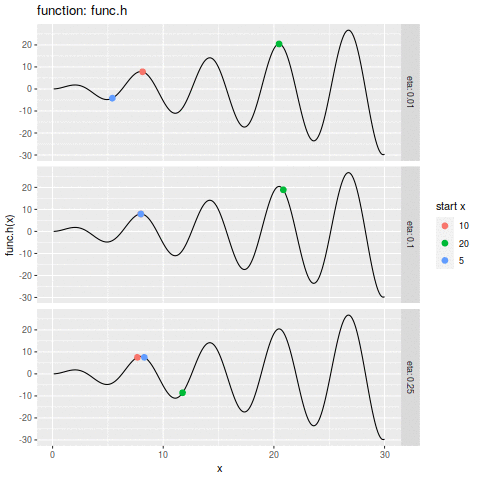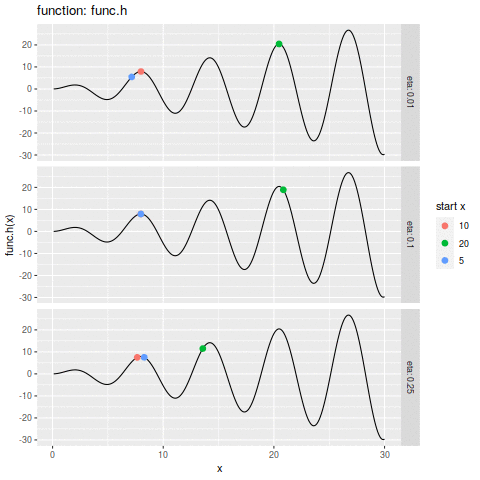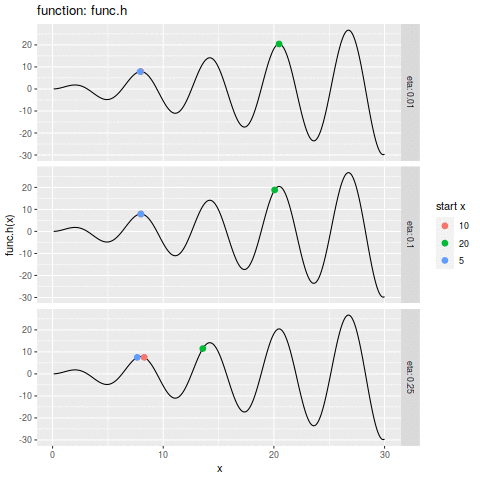
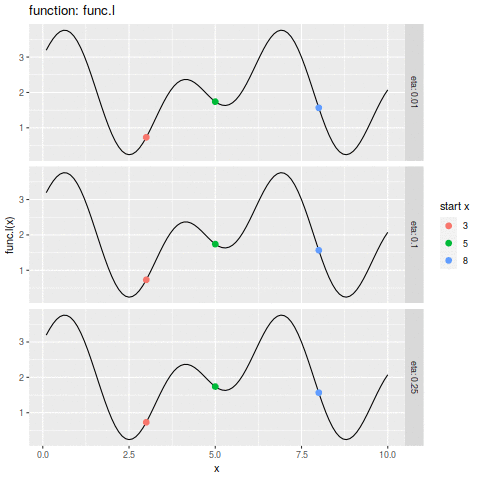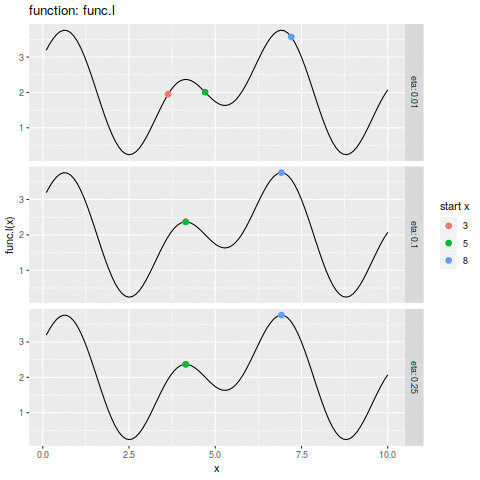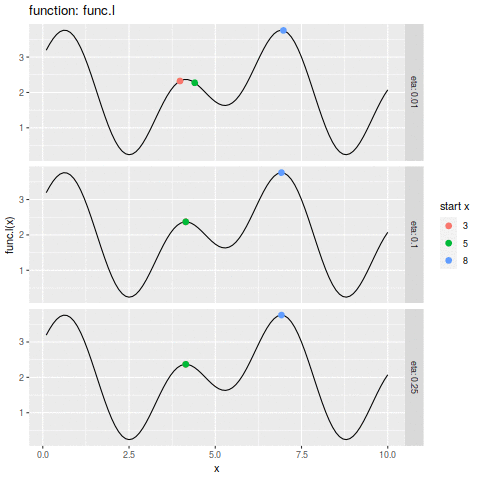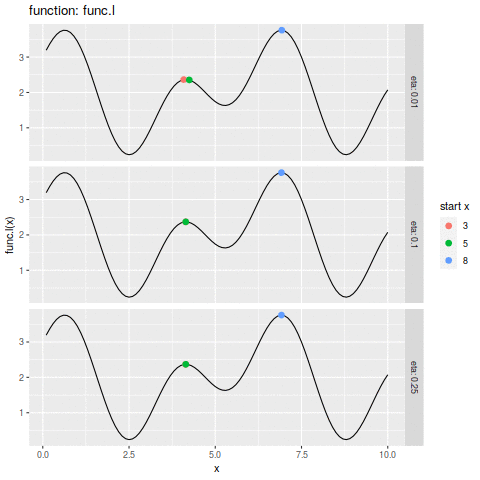
commit
ca06e3526a
32 changed files with 666 additions and 0 deletions
Split View
Diff Options
-
+2 -0.gitignore
-
BINue01/cpu_times.pdf
-
+11 -0ue01/sum1.R
-
+10 -0ue01/sum2.R
-
+13 -0ue01/sum2.c
-
BINue01/sum2.o
-
BINue01/sum2.so
-
+9 -0ue01/test1.R
-
+20 -0ue01/time_sum.R
-
+9 -0ue01/twosums/DESCRIPTION
-
+1 -0ue01/twosums/NAMESPACE
-
+12 -0ue01/twosums/R/sum1.R
-
+11 -0ue01/twosums/R/sum2.R
-
+10 -0ue01/twosums/Read-and-delete-me
-
+69 -0ue01/twosums/man/sum1.Rd
-
+65 -0ue01/twosums/man/sum2.Rd
-
+36 -0ue01/twosums/man/twosums-package.Rd
-
+13 -0ue01/twosums/src/sum2.c
-
BINue01/ue01.zip
-
BINue02/a1/a/func_g.gif
-
BINue02/a1/a/func_g.pdf
-
BINue02/a1/a/func_h.gif
-
BINue02/a1/a/func_h.pdf
-
BINue02/a1/a/func_k.gif
-
BINue02/a1/a/func_k.pdf
-
BINue02/a1/a/func_l.gif
-
BINue02/a1/a/func_l.pdf
-
+126 -0ue02/a1/a/gradient_ascent.R
-
+124 -0ue02/a1/b/2
-
+125 -0ue02/a1/b/ea_alg.R
-
BINue02/a1/b/plot.pdf
-
+0 -0ue02/a2/ea_crossover.R
+ 2
- 0
.gitignore
View File
| @ -0,0 +1,2 @@ | |||
| .RData | |||
| .Rhistory | |||
BIN
ue01/cpu_times.pdf
View File
+ 11
- 0
ue01/sum1.R
View File
| @ -0,0 +1,11 @@ | |||
| sum1 <- function(m) { | |||
| res = 0 | |||
| for(i in 1:dim(m)[1]) { | |||
| for(j in 1:dim(m)[2]) { | |||
| res <- res + m[i,j] | |||
| } | |||
| } | |||
| return(res) | |||
| } | |||
+ 10
- 0
ue01/sum2.R
View File
| @ -0,0 +1,10 @@ | |||
| sum2 <- function(m) { | |||
| res = .C("sum2" | |||
| ,mtrx=as.double(m) | |||
| ,m=as.integer(dim(m)[1]) | |||
| ,n=as.integer(dim(m)[2]) | |||
| ,res=as.double(0) | |||
| ) | |||
| return(res$res) | |||
| } | |||
+ 13
- 0
ue01/sum2.c
View File
| @ -0,0 +1,13 @@ | |||
| #include <R.h> | |||
| void sum2(double* mtrx, int* m, int* n, double* res) | |||
| { | |||
| *res = 0; | |||
| for(int i=0; i < *n; i++) | |||
| { | |||
| for(int j=0; j < *m; j++) { | |||
| *res += mtrx[j * (*n) + i]; | |||
| } | |||
| } | |||
| } | |||
BIN
ue01/sum2.o
View File
BIN
ue01/sum2.so
View File
+ 9
- 0
ue01/test1.R
View File
| @ -0,0 +1,9 @@ | |||
| # test1.R | |||
| x <- c(1,2,5,7) | |||
| mx <- mean(x) # Mittelwert berechnen | |||
| sx <- sd(x) # Standardabweichung berechnen | |||
| cat("x = ", x, "\n") | |||
| cat("Mittelwert = ", mx, "\n") | |||
| cat("Standardabweichung = ", sx, "\n") | |||
+ 20
- 0
ue01/time_sum.R
View File
| @ -0,0 +1,20 @@ | |||
| source("sum1.R") | |||
| source("sum2.R") | |||
| dyn.load("sum2.so") | |||
| comp_sum_funcs <- function(sizes){ | |||
| msizes = c() | |||
| times_1 = c() | |||
| times_2 = c() | |||
| for(i in sizes) { | |||
| m <- matrix(rnorm(i^2), i) | |||
| times_1 = append(times_1, summary(system.time(sum1(m)))[1]) | |||
| times_2 = append(times_2, summary(system.time(sum2(m)))[1]) | |||
| msizes = append(msizes, i) | |||
| } | |||
| return(data.frame(size=msizes, sum_1 = times_1, sum_2 = times_2)) | |||
| } | |||
+ 9
- 0
ue01/twosums/DESCRIPTION
View File
| @ -0,0 +1,9 @@ | |||
| Package: twosums | |||
| Type: Package | |||
| Title: What the package does (short line) | |||
| Version: 1.0 | |||
| Date: 2021-05-12 | |||
| Author: Who wrote it | |||
| Maintainer: Who to complain to <yourfault@somewhere.net> | |||
| Description: More about what it does (maybe more than one line) | |||
| License: What license is it under? | |||
+ 1
- 0
ue01/twosums/NAMESPACE
View File
| @ -0,0 +1 @@ | |||
| export("sum1", "sum2") | |||
+ 12
- 0
ue01/twosums/R/sum1.R
View File
| @ -0,0 +1,12 @@ | |||
| sum1 <- | |||
| function(m) { | |||
| res = 0 | |||
| for(i in 1:dim(m)[1]) { | |||
| for(j in 1:dim(m)[2]) { | |||
| res <- res + m[i,j] | |||
| } | |||
| } | |||
| return(res) | |||
| } | |||
+ 11
- 0
ue01/twosums/R/sum2.R
View File
| @ -0,0 +1,11 @@ | |||
| sum2 <- | |||
| function(m) { | |||
| res = .C("sum2" | |||
| ,mtrx=as.double(m) | |||
| ,m=as.integer(dim(m)[1]) | |||
| ,n=as.integer(dim(m)[2]) | |||
| ,res=as.double(0) | |||
| ) | |||
| return(res$res) | |||
| } | |||
+ 10
- 0
ue01/twosums/Read-and-delete-me
View File
| @ -0,0 +1,10 @@ | |||
| * Edit the help file skeletons in 'man', possibly combining help files | |||
| for multiple functions. | |||
| * Edit the exports in 'NAMESPACE', and add necessary imports. | |||
| * Put any C/C++/Fortran code in 'src'. | |||
| * If you have compiled code, add a useDynLib() directive to | |||
| 'NAMESPACE'. | |||
| * Run R CMD build to build the package tarball. | |||
| * Run R CMD check to check the package tarball. | |||
| Read "Writing R Extensions" for more information. | |||
+ 69
- 0
ue01/twosums/man/sum1.Rd
View File
| @ -0,0 +1,69 @@ | |||
| \name{sum1} | |||
| \alias{sum1} | |||
| %- Also NEED an '\alias' for EACH other topic documented here. | |||
| \title{ | |||
| %% ~~function to do ... ~~ | |||
| } | |||
| \description{ | |||
| %% ~~ A concise (1-5 lines) description of what the function does. ~~ | |||
| } | |||
| \usage{ | |||
| sum1(m) | |||
| } | |||
| %- maybe also 'usage' for other objects documented here. | |||
| \arguments{ | |||
| \item{m}{ | |||
| %% ~~Describe \code{m} here~~ | |||
| } | |||
| } | |||
| \details{ | |||
| %% ~~ If necessary, more details than the description above ~~ | |||
| } | |||
| \value{ | |||
| %% ~Describe the value returned | |||
| %% If it is a LIST, use | |||
| %% \item{comp1 }{Description of 'comp1'} | |||
| %% \item{comp2 }{Description of 'comp2'} | |||
| %% ... | |||
| } | |||
| \references{ | |||
| %% ~put references to the literature/web site here ~ | |||
| } | |||
| \author{ | |||
| %% ~~who you are~~ | |||
| } | |||
| \note{ | |||
| %% ~~further notes~~ | |||
| } | |||
| %% ~Make other sections like Warning with \section{Warning }{....} ~ | |||
| \seealso{ | |||
| %% ~~objects to See Also as \code{\link{help}}, ~~~ | |||
| } | |||
| \examples{ | |||
| ##---- Should be DIRECTLY executable !! ---- | |||
| ##-- ==> Define data, use random, | |||
| ##-- or do help(data=index) for the standard data sets. | |||
| ## The function is currently defined as | |||
| function (m) | |||
| { | |||
| res = 0 | |||
| for (i in 1:dim(m)[1]) { | |||
| for (j in 1:dim(m)[2]) { | |||
| res <- res + m[i, j] | |||
| } | |||
| } | |||
| return(res) | |||
| } | |||
| } | |||
| % Add one or more standard keywords, see file 'KEYWORDS' in the | |||
| % R documentation directory (show via RShowDoc("KEYWORDS")): | |||
| % \keyword{ ~kwd1 } | |||
| % \keyword{ ~kwd2 } | |||
| % Use only one keyword per line. | |||
| % For non-standard keywords, use \concept instead of \keyword: | |||
| % \concept{ ~cpt1 } | |||
| % \concept{ ~cpt2 } | |||
| % Use only one concept per line. | |||
+ 65
- 0
ue01/twosums/man/sum2.Rd
View File
| @ -0,0 +1,65 @@ | |||
| \name{sum2} | |||
| \alias{sum2} | |||
| %- Also NEED an '\alias' for EACH other topic documented here. | |||
| \title{ | |||
| %% ~~function to do ... ~~ | |||
| } | |||
| \description{ | |||
| %% ~~ A concise (1-5 lines) description of what the function does. ~~ | |||
| } | |||
| \usage{ | |||
| sum2(m) | |||
| } | |||
| %- maybe also 'usage' for other objects documented here. | |||
| \arguments{ | |||
| \item{m}{ | |||
| %% ~~Describe \code{m} here~~ | |||
| } | |||
| } | |||
| \details{ | |||
| %% ~~ If necessary, more details than the description above ~~ | |||
| } | |||
| \value{ | |||
| %% ~Describe the value returned | |||
| %% If it is a LIST, use | |||
| %% \item{comp1 }{Description of 'comp1'} | |||
| %% \item{comp2 }{Description of 'comp2'} | |||
| %% ... | |||
| } | |||
| \references{ | |||
| %% ~put references to the literature/web site here ~ | |||
| } | |||
| \author{ | |||
| %% ~~who you are~~ | |||
| } | |||
| \note{ | |||
| %% ~~further notes~~ | |||
| } | |||
| %% ~Make other sections like Warning with \section{Warning }{....} ~ | |||
| \seealso{ | |||
| %% ~~objects to See Also as \code{\link{help}}, ~~~ | |||
| } | |||
| \examples{ | |||
| ##---- Should be DIRECTLY executable !! ---- | |||
| ##-- ==> Define data, use random, | |||
| ##-- or do help(data=index) for the standard data sets. | |||
| ## The function is currently defined as | |||
| function (m) | |||
| { | |||
| res = .C("sum2", mtrx = as.double(m), m = as.integer(dim(m)[1]), | |||
| n = as.integer(dim(m)[2]), res = as.double(0)) | |||
| return(res$res) | |||
| } | |||
| } | |||
| % Add one or more standard keywords, see file 'KEYWORDS' in the | |||
| % R documentation directory (show via RShowDoc("KEYWORDS")): | |||
| % \keyword{ ~kwd1 } | |||
| % \keyword{ ~kwd2 } | |||
| % Use only one keyword per line. | |||
| % For non-standard keywords, use \concept instead of \keyword: | |||
| % \concept{ ~cpt1 } | |||
| % \concept{ ~cpt2 } | |||
| % Use only one concept per line. | |||
+ 36
- 0
ue01/twosums/man/twosums-package.Rd
View File
| @ -0,0 +1,36 @@ | |||
| \name{twosums-package} | |||
| \alias{twosums-package} | |||
| \alias{twosums} | |||
| \docType{package} | |||
| \title{ | |||
| \packageTitle{twosums} | |||
| } | |||
| \description{ | |||
| \packageDescription{twosums} | |||
| } | |||
| \details{ | |||
| The DESCRIPTION file: | |||
| \packageDESCRIPTION{twosums} | |||
| \packageIndices{twosums} | |||
| ~~ An overview of how to use the package, including the most important ~~ | |||
| ~~ functions ~~ | |||
| } | |||
| \author{ | |||
| \packageAuthor{twosums} | |||
| Maintainer: \packageMaintainer{twosums} | |||
| } | |||
| \references{ | |||
| ~~ Literature or other references for background information ~~ | |||
| } | |||
| ~~ Optionally other standard keywords, one per line, from file KEYWORDS in ~~ | |||
| ~~ the R documentation directory ~~ | |||
| \keyword{ package } | |||
| \seealso{ | |||
| ~~ Optional links to other man pages, e.g. ~~ | |||
| ~~ \code{\link[<pkg>:<pkg>-package]{<pkg>}} ~~ | |||
| } | |||
| \examples{ | |||
| ~~ simple examples of the most important functions ~~ | |||
| } | |||
+ 13
- 0
ue01/twosums/src/sum2.c
View File
| @ -0,0 +1,13 @@ | |||
| #include <R.h> | |||
| void sum2(double* mtrx, int* m, int* n, double* res) | |||
| { | |||
| *res = 0; | |||
| for(int i=0; i < *n; i++) | |||
| { | |||
| for(int j=0; j < *m; j++) { | |||
| *res += mtrx[j * (*n) + i]; | |||
| } | |||
| } | |||
| } | |||
BIN
ue01/ue01.zip
View File
BIN
ue02/a1/a/func_g.gif
View File
BIN
ue02/a1/a/func_g.pdf
View File
BIN
ue02/a1/a/func_h.gif
View File
BIN
ue02/a1/a/func_h.pdf
View File
BIN
ue02/a1/a/func_k.gif
View File
BIN
ue02/a1/a/func_k.pdf
View File
BIN
ue02/a1/a/func_l.gif
View File
BIN
ue02/a1/a/func_l.pdf
View File
+ 126
- 0
ue02/a1/a/gradient_ascent.R
View File
| @ -0,0 +1,126 @@ | |||
| library(ggplot2) | |||
| library(gganimate) | |||
| library(patchwork) | |||
| gradient <- function(f, x, d){ | |||
| return((f(x + d) - f(x - d)) / (2*d)) | |||
| } | |||
| gradient.ascent.move <- function(f, x, d, mu){ | |||
| return(x + mu * gradient(f, x, d)) | |||
| } | |||
| func.g <- function(x) x | |||
| func.k <- function(x) sin(x) | |||
| func.h <- function(x) x * sin(x) | |||
| func.l <- function(x) 2 + cos(x) + sin(2*x) | |||
| gradient.ascent.iterate <- function(f, x, d, mu, n){ | |||
| if(n == 1) { | |||
| return(gradient.ascent.move(f, x, d, mu)) | |||
| } | |||
| return(gradient.ascent.niter(f | |||
| ,gradient.descent.move(f, x, d, mu) | |||
| ,d | |||
| ,mu | |||
| ,n-1 | |||
| )) | |||
| } | |||
| gradient.ascent.iterverb <- function(f, x, d, mu, n, xs=numeric()){ | |||
| next_x <- gradient.ascent.move(f, x, d, mu) | |||
| xs[length(xs)+1] <- next_x | |||
| if(n == 1) { | |||
| return(xs) | |||
| } | |||
| return(gradient.ascent.iterverb(f, next_x, d, mu, n-1, xs)) | |||
| } | |||
| trace.ascent <- function(f, x, d, eta, n, xs) { | |||
| df_dc = data.frame(x=numeric() | |||
| ,y=numeric() | |||
| ,i=integer() | |||
| ,start_x=character() | |||
| ,eta=numeric()) | |||
| for(start in x) { | |||
| for(e in eta) { | |||
| first_it <- TRUE | |||
| if(first_it == TRUE) { | |||
| df_dc <- rbind(df_dc, data.frame(x=c(start) | |||
| ,y=c(f(start)) | |||
| ,i=c(0) | |||
| ,start_x=c(as.character(start)) | |||
| ,eta=c(e) | |||
| )) | |||
| first_it <- FALSE | |||
| } | |||
| xf <- gradient.ascent.iterverb(f, start, d, e, n) | |||
| df_dc <- rbind(df_dc, data.frame(x=xf | |||
| ,y=f(xf) | |||
| ,i=1:length(xf) | |||
| ,start_x=rep(as.character(start), length(xf)) | |||
| ,eta=e) | |||
| ) | |||
| } | |||
| } | |||
| return(df_dc) | |||
| } | |||
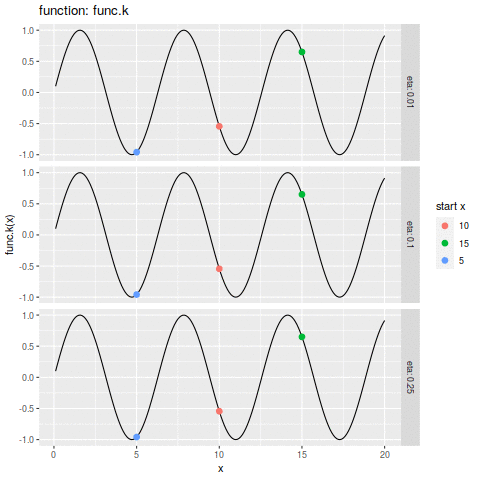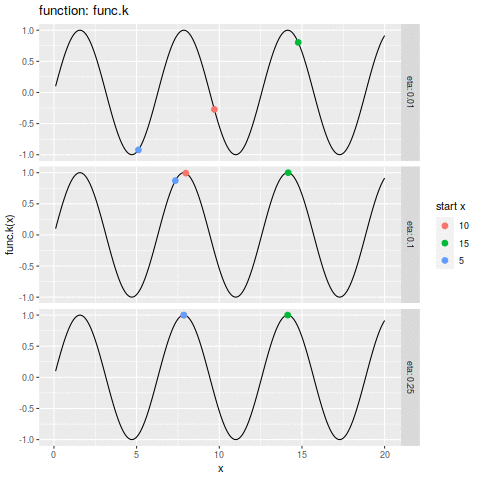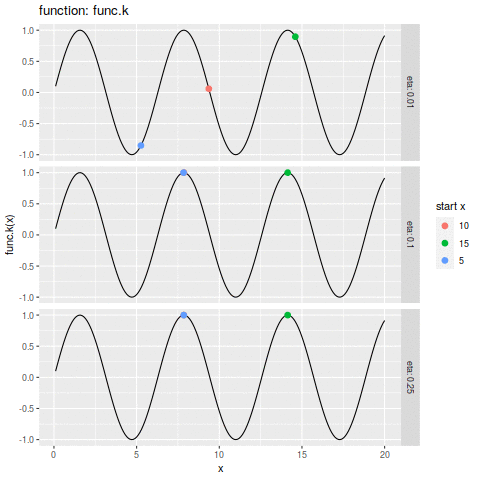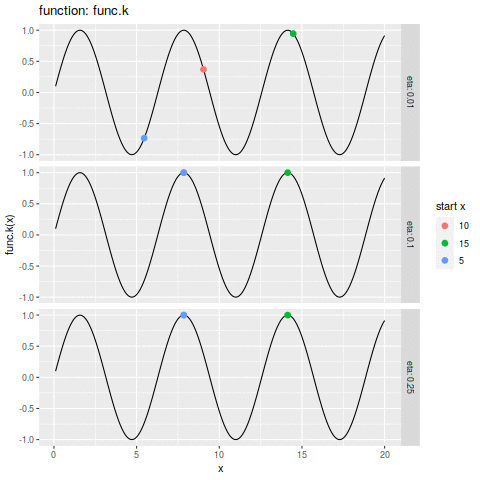
| plot.ascent <- function(f, x, d, eta, n, xs) { | |||
| df_dc = trace.ascent(f, x, d, eta, n, xs) | |||
| func_str = deparse(substitute(f)) | |||
| df_f <- data.frame(x=xs, y=f(xs)) | |||
| p1 <- ggplot(df_f, aes(x=x, y=y)) + | |||
| geom_line() + | |||
| geom_point(aes(colour=start_x | |||
| ,size=i | |||
| ) | |||
| ,data=df_dc) + | |||
| labs(size="iteration" | |||
| ,alpha="iteration" | |||
| ,color="start x" | |||
| ,y=sprintf("%s(x)", func_str)) + | |||
| facet_grid(eta ~ ., labeller=label_both) | |||
| p2 <- ggplot(df_dc, aes(x=i, y=y)) + | |||
| geom_line(aes(colour=start_x), show.legend=FALSE) + | |||
| labs(x="iteration" | |||
| ,y=sprintf("%s(x)", func_str)) + | |||
| facet_grid(eta ~ ., labeller=label_both) | |||
| p <- (p1 | p2) + | |||
| plot_annotation(title=sprintf("function: %s", func_str)) + | |||
| plot_layout(guides="collect" | |||
| ,widths=10 | |||
| ,heights=2) | |||
| return(p) | |||
| } | |||
| animate.ascent <- function(f, x, d, eta, n, xs) { | |||
| df_dc = trace.ascent(f, x, d, eta, n, xs) | |||
| func_str <- deparse(substitute(f)) | |||
| df_f <- data.frame(x=xs, y=f(xs)) | |||
| p <- ggplot(df_f, aes(x=x, y=y)) + | |||
| geom_line() + | |||
| geom_point(aes(colour=start_x), size=2.5, data=df_dc) + | |||
| labs(color="start x", y=sprintf("%s(x)", func_str)) + | |||
| facet_grid(eta ~ ., labeller=label_both) + | |||
| ggtitle(sprintf("function: %s", func_str)) | |||
| anim <- p + transition_reveal(i) | |||
| return(anim) | |||
| } | |||
+ 124
- 0
ue02/a1/b/2
View File
| @ -0,0 +1,124 @@ | |||
| library(ggplot2) | |||
| func.g <- function(x) x | |||
| func.k <- function(x) sin(x) | |||
| func.h <- function(x) x * sin(x) | |||
| func.l <- function(x) 2 + cos(x) + sin(2*x) | |||
| delta.const <- function(x) { | |||
| return(function() x) | |||
| } | |||
| delta.gaus <- function() { | |||
| return(rnorm(1)) | |||
| } | |||
| ea.init_population <- function(range, size, rand_gen) { | |||
| return(rand_gen(size) * (range[2] - range[1]) + range[1]) | |||
| } | |||
| ea.trace <- function(range, delta, population_size, fit_func, iterations) { | |||
| population <- ea.init_population(range, population_size, runif) | |||
| df <- data.frame(i=integer() | |||
| ,max=numeric() | |||
| ,median=numeric() | |||
| ,min=numeric()) | |||
| for(i in 1:iterations) { | |||
| population <- ea.iterate(delta, population, fit_func) | |||
| df[nrow(df) + 1,] <- c(i | |||
| ,population[1] | |||
| ,population[length(population) %/% 2] | |||
| ,population[length(population)] | |||
| ) | |||
| } | |||
| df["max_val"] <- fit_func(df$max) | |||
| df["median_val"] <- fit_func(df$median) | |||
| df["min_val"] <- fit_func(df$min) | |||
| res <- list(population, df) | |||
| names(res) <- c("population", "df_tr") | |||
| return(res) | |||
| } | |||
| ea.traces <- function(range, deltas, population_size, fit_funcs, iterations) { | |||
| df <- data.frame(i=integer() | |||
| ,max=numeric() | |||
| ,median=numeric() | |||
| ,min=numeric() | |||
| ,delta_func=character() | |||
| ,delta=numeric() | |||
| ,fit_func=character()) | |||
| for(delta_func in names(deltas)) { | |||
| delta <- deltas[delta_func] | |||
| for(fit_name in names(fit_funcs)) { | |||
| fit <- fit_funcs[fit_name] | |||
| tmp_df <- ea.trace(range, delta, population_size, fit, iterations)$df_tr | |||
| tmp_df["delta_func"] <- delta_func | |||
| if(delta_func == "delta.gaus") { | |||
| tmp_df["delta"] <- NA | |||
| } else { | |||
| tmp_df["delta"] <- delta() | |||
| } | |||
| tmp_df["fit_func"] <- fit_name | |||
| df <- rbind(df, tmp_df) | |||
| } | |||
| } | |||
| return(df) | |||
| } | |||
| ea.plot <- function(range, delta, population_size, fit_func, iterations) { | |||
| res <- ea.trace(range, delta, population_size, fit_func, iterations) | |||
| df_vals <- melt(res$df_tr[c("i", "min_val", "median_val", "max_val")], id.vars="i") | |||
| p <- ggplot(data=df_vals, aes(x=i)) + | |||
| geom_line(aes(y=value, linetype=variable)) | |||
| return(p) | |||
| } | |||
| ea.run <- function(range, delta, population_size, fit_func, iterations) { | |||
| population <- ea.init_population(range, population_size, runif) | |||
| for(i in 1:iterations) { | |||
| population <- ea.iterate(delta, population, fit_func) | |||
| } | |||
| return(population) | |||
| } | |||
| ea.iterate <- function(delta, population, fit_func) { | |||
| children <- c() | |||
| for(individual in population) { | |||
| children <- append(children, ea.mutate(individual, delta)) | |||
| } | |||
| population <- append(population, children) | |||
| return(ea.select(population, fit_func)) | |||
| } | |||
| ea.mutate <- function(individual, delta) { | |||
| sign <- sample(c(-1,1), 1) | |||
| return(individual + sign * delta()) | |||
| } | |||
| ea.select <- function(population, fit_func) { | |||
| sorted_popul <- population[order(sapply(population, fit_func), decreasing=TRUE)] | |||
| return(sorted_popul[1 : (length(sorted_popul) %/% 2)]) | |||
| } | |||
+ 125
- 0
ue02/a1/b/ea_alg.R
View File
| @ -0,0 +1,125 @@ | |||
| library(ggplot2) | |||
| func.g <- function(x) x | |||
| func.k <- function(x) sin(x) | |||
| func.h <- function(x) x * sin(x) | |||
| func.l <- function(x) 2 + cos(x) + sin(2*x) | |||
| delta.const <- function(x) { | |||
| return(function() x) | |||
| } | |||
| delta.gaus <- function() { | |||
| return(rnorm(1)) | |||
| } | |||
| ea.init_population <- function(range, size, rand_gen) { | |||
| return(rand_gen(size) * (range[2] - range[1]) + range[1]) | |||
| } | |||
| ea.trace <- function(range, delta, population_size, fit_func, iterations) { | |||
| population <- ea.init_population(range, population_size, runif) | |||
| df <- data.frame(i=integer() | |||
| ,max=numeric() | |||
| ,median=numeric() | |||
| ,min=numeric()) | |||
| for(i in 1:iterations) { | |||
| population <- ea.iterate(delta, population, fit_func) | |||
| df[nrow(df) + 1,] <- c(i | |||
| ,population[1] | |||
| ,population[length(population) %/% 2] | |||
| ,population[length(population)] | |||
| ) | |||
| } | |||
| df["max_val"] <- fit_func(df$max) | |||
| df["median_val"] <- fit_func(df$median) | |||
| df["min_val"] <- fit_func(df$min) | |||
| res <- list(population, df) | |||
| names(res) <- c("population", "df_tr") | |||
| return(res) | |||
| } | |||
| ea.traces <- function(range, deltas, population_size, fit_funcs, iterations) { | |||
| df <- data.frame(i=integer() | |||
| ,max=numeric() | |||
| ,median=numeric() | |||
| ,min=numeric() | |||
| ,delta_func=character() | |||
| ,delta=numeric() | |||
| ,fit_func=character()) | |||
| for(delta_func in names(deltas)) { | |||
| delta <- deltas[[delta_func]] | |||
| for(fit_name in names(fit_funcs)) { | |||
| fit <- fit_funcs[[fit_name]] | |||
| print(delta_func) | |||
| tmp_df <- ea.trace(range, delta, population_size, fit, iterations)$df_tr | |||
| tmp_df["delta_func"] <- delta_func | |||
| if(delta_func == "delta.gaus") { | |||
| tmp_df["delta"] <- NA | |||
| } else { | |||
| tmp_df["delta"] <- delta() | |||
| } | |||
| tmp_df["fit_func"] <- fit_name | |||
| df <- rbind(df, tmp_df) | |||
| } | |||
| } | |||
| return(df) | |||
| } | |||
| ea.plot <- function(range, delta, population_size, fit_func, iterations) { | |||
| res <- ea.trace(range, delta, population_size, fit_func, iterations) | |||
| df_vals <- melt(res$df_tr[c("i", "min_val", "median_val", "max_val")], id.vars="i") | |||
| p <- ggplot(data=df_vals, aes(x=i)) + | |||
| geom_line(aes(y=value, linetype=variable)) | |||
| return(p) | |||
| } | |||
| ea.run <- function(range, delta, population_size, fit_func, iterations) { | |||
| population <- ea.init_population(range, population_size, runif) | |||
| for(i in 1:iterations) { | |||
| population <- ea.iterate(delta, population, fit_func) | |||
| } | |||
| return(population) | |||
| } | |||
| ea.iterate <- function(delta, population, fit_func) { | |||
| children <- c() | |||
| for(individual in population) { | |||
| children <- append(children, ea.mutate(individual, delta)) | |||
| } | |||
| population <- append(population, children) | |||
| return(ea.select(population, fit_func)) | |||
| } | |||
| ea.mutate <- function(individual, delta) { | |||
| sign <- sample(c(-1,1), 1) | |||
| return(individual + sign * delta()) | |||
| } | |||
| ea.select <- function(population, fit_func) { | |||
| sorted_popul <- population[order(sapply(population, fit_func), decreasing=TRUE)] | |||
| return(sorted_popul[1 : (length(sorted_popul) %/% 2)]) | |||
| } | |||